☰ Choose Image
Dimensions (X x Y x Z x C x T)
$ build/bioformats2raw-0.2.1/bin/bioformats2raw \
--file_type=zarr \
--dimension-order=XYZCT \
a.fake \
output
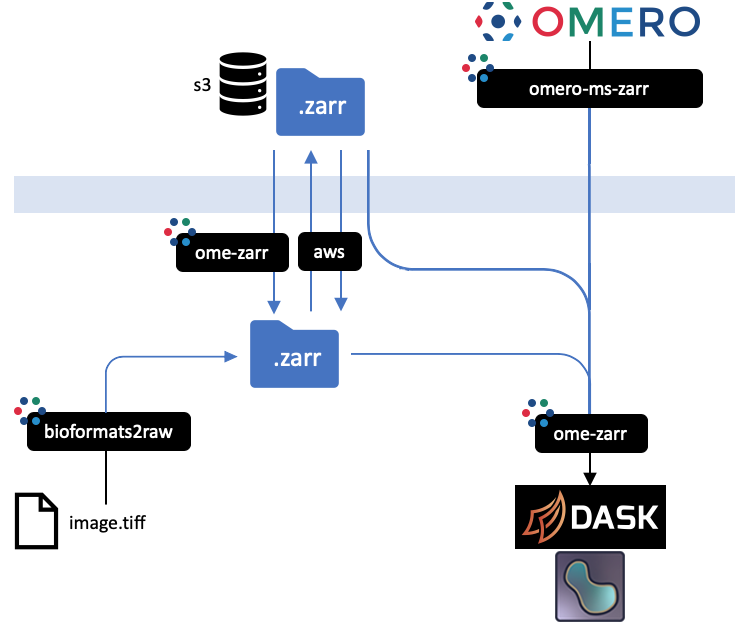
$ find output
output
output/METADATA.ome.xml
output/data.zarr
output/data.zarr/.zattrs
output/data.zarr/.zgroup
output/data.zarr/0
output/data.zarr/0/.zattrs
output/data.zarr/0/.zgroup
output/data.zarr/0/0
output/data.zarr/0/0/0.0.0.0.0
output/data.zarr/0/0/.zarray
output/data.zarr/0/1
output/data.zarr/0/1/0.0.0.0.0
output/data.zarr/0/1/.zarray
$ mv output/data.zarr/0 /data/v0.1/6001240.zarr
$ pip install awscli
# upload
aws --endpoint-url https://s3.embassy.ebi.ac.uk cp \
--acl public-read --recursive \
/data/v0.1/6001240.zarr/ \
s3://idr/zarr/v0.1/6001240.zarr
# download
$ aws --endpoint-url --no-sign-request \
https://s3.embassy.ebi.ac.uk \
s3 cp --recursive s3://idr/zarr/v0.1/6001240.zarr \
/tmp/
$ git clone -b v0.1.1 https://github.com/ome/omero-ms-zarr
$ gradle build -x test
$ cat <<EOF > cfg
omero.data.dir=/tmp/ome1
omero.db.name=ome1
omero.db.user=postgres
omero.ms.zarr.net.path.image=/idr/zarr/v0.1/6001240.zarr/
EOF
$ gradle run --args=cfg
# https://workshop.openmicroscopy.org/image/6001240.zarr
$ pip install ome-zarr
$ ome_zarr download https://s3.embassy.ebi.ac.uk/6001240.zarr
$ ome_zarr info 6001240.zarr
url = 'https://s3.embassy.ebi.ac.uk/6001240.zarr'
zarr = ome_zarr.parse_url(url)
zarr.is_ome_zarr()
data = zarr.load_ome_zarr()
pyramid = data[0]
$ napari 'https://s3.embassy.ebi.ac.uk/6001240.zarr'
from dask.diagnostics import ProgressBar
import dask.array as da
import numpy as np
with ProgressBar():
array = "https://s3.embassy.ebi.ac.uk/6001240.zarr/1"
print(np.asarray(da.from_zarr(array)))
[########################################] | 100% Completed | 16.5s
[[[[[ 8 8 8 ... 16 15 9]
[ 8 8 9 ... 13 9 26]
[ 9 8 9 ... 9 8 8]
...
...