What type of data?
- New light sheet microscopy techniques
- Large volumetric datasets (terabyte-sized)
- Increased numbers of dimensions and scale
- Multi view datasets
- Performance limitations with TIFF based formats
- Need for fast random data access across dimensions
- New formats developed by the community
- Images stored as chunked multidimensional arrays
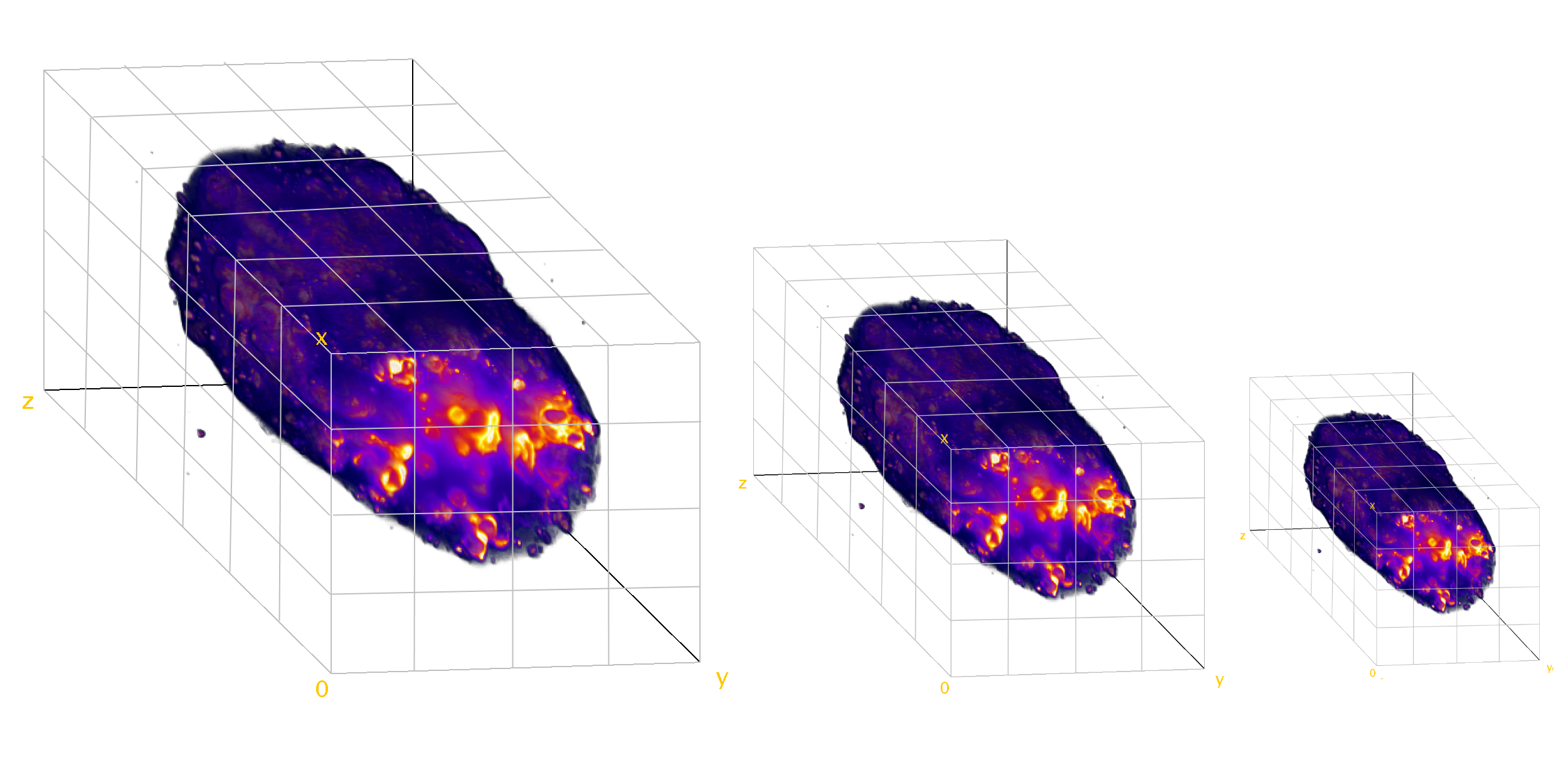

Current Scenario?
OME-TIFF
- A number of existing formats such as Imaris and CellH5
- These are often HDF5 based
- Images are converted to OME-TIFF format
- Not as performant as desired when scale increases
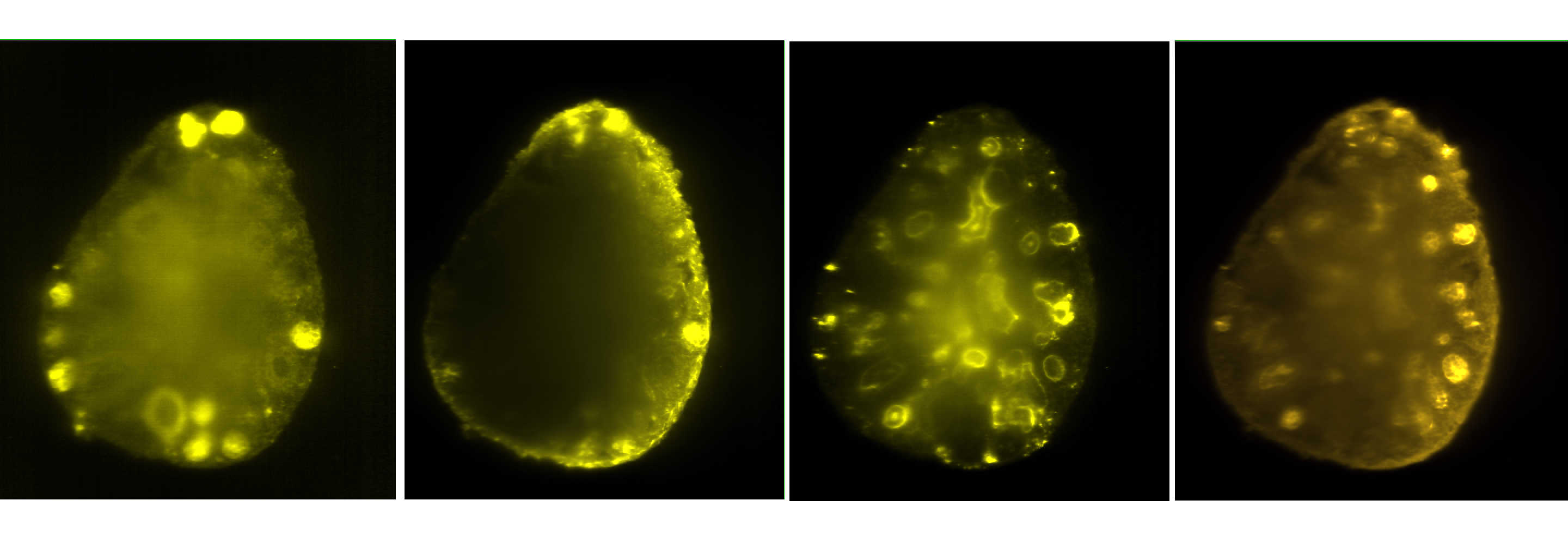
OME-Model
- Metadata stored in 5D plane-based model
- A new image and a new Pixels element for each image volume
- Additional metadata stored as Annotations
Bio-Formats API
- Original API allowed for reading and writing of planes
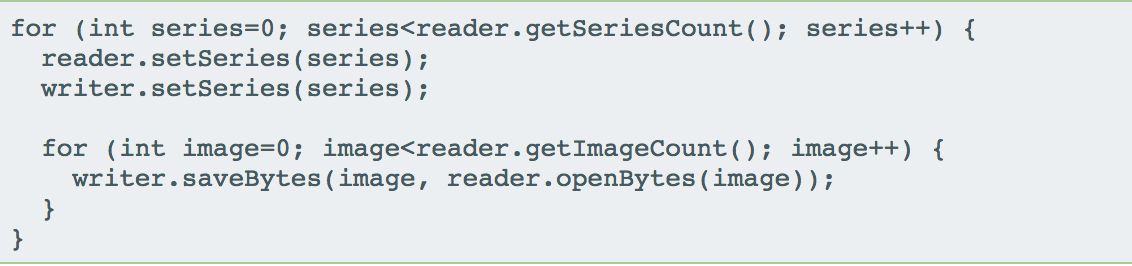
- Last year we had API additions to support tiling

What is the solution?
- New binary container format to sit alongside OME-TIFF
- New OME model which supports nD chunk-based data
- Full backwards compatibility
- New API for reading and writing of chunked data
- A way to represent multiple volumes as a single image
- The ability to read and write multi dimensional images
- Flexibility to add user defined dimensions and attributes
What have we done so far?
Existing OME model
- Consists of a single Pixels element for every Image
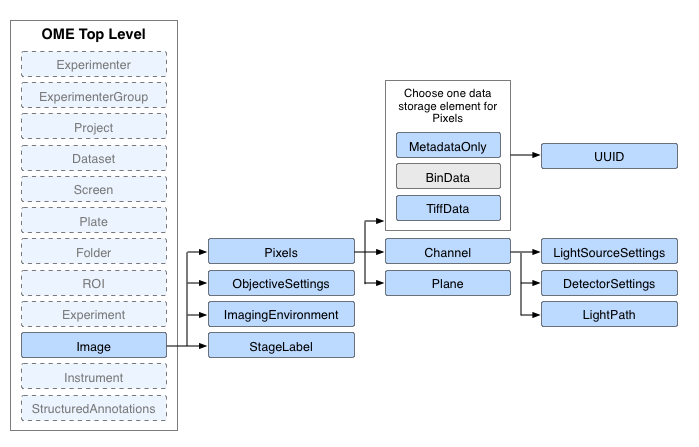
Existing Pixels element
- 5 dimensions - X, Y, Z, T and C
- Multiple Channel elements per Pixels
- TiffData stores IFD
- Pixels element for each image volume
Proposed OME model
- Experimental - still in early scoping phases
- Conversion of the existing 5D plane-based model to an nD chunk-based model
- The ability and flexibility to add user defined dimensions and attributes
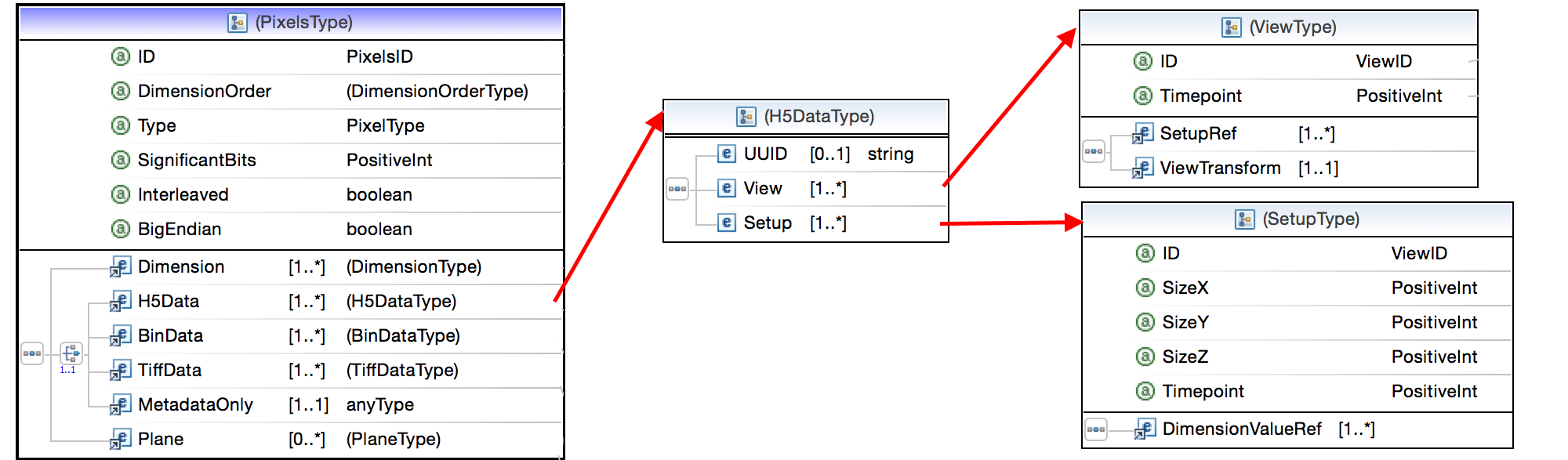
Example OME-XML

Big Data Viewer (BDV)
- The BigDataViewer from MPI Dresden
- A re-slicing browser in Fiji's SPIMage processing pipeline
- Also has a custom data format based on XML and HDF5
- Images are tiled multi-resolution pyramids
- Each channel or angle or combination of both is a setup
- Each combination of setup and timepoint as a view.
- Each view has one corresponding image volume.
- https://imagej.net/BigDataViewer
Keller Lab Block file type (KLB)
Our plans for the future
- Integrate the new formats into the existing OME-Model
- Continue scoping new OME container
- Prototyping and testing new iterations of the data model
- New API additions to support extended dimensions
- Design new API additions for chunk based access
- Develop new container format
- Implement reader and writer for new OME format















