OMERO workshop
TOPIM TECH
Chania, July 2018
Will Moore, Petr Walczysko
Today's workshop
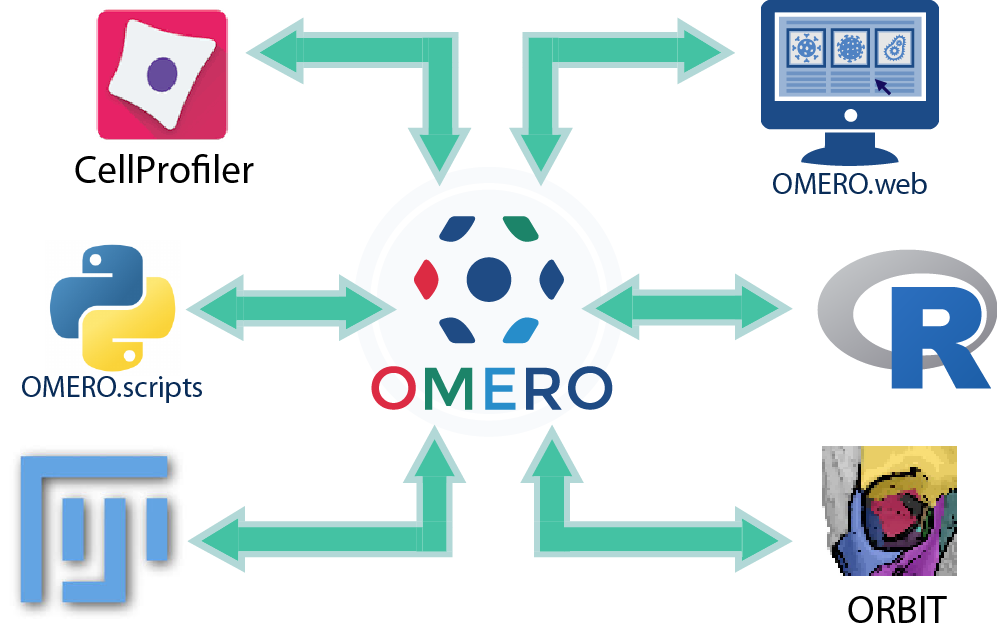
Outline

- 11:20 - 12:55 (1 hour 35 mins)
- Introduction to OMERO
- CellProfiler analysis
- 13:25 - 14:10 (45 mins)
- Image analysis in Orbit
- Coding in Orbit
- Lunch
- 18:00 - 18:45 (45 mins)
- Fiji analysis
- Python script
- R analysis
- 19:10 - 20:15 (1 hour 5 mins)
- OMERO.parade
- OMERO.figure
Client-side vs Server-side analysis
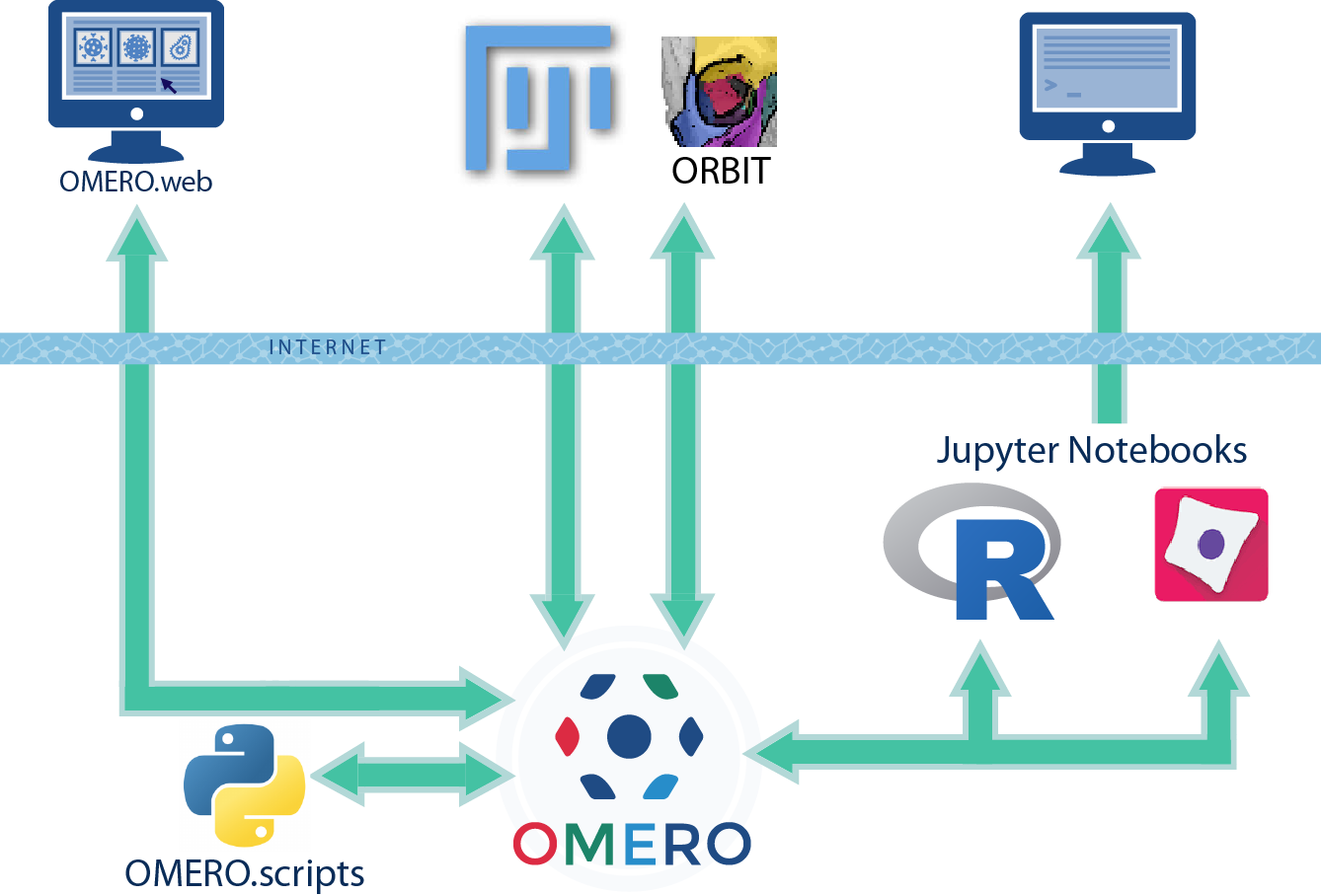
Analysis data in OMERO
- OMERO.tables
- HDF tables
- 1 row per Image / Well - flexible columns
- Saved as a File-Annotation
-
Regions of Interest (ROIs)
- Contain 1 or more Shape e.g. Polygon
-
Map Annotations
- List of Key: Value pairs
- Each map annotation linked to 1 object
Session 1: OMERO intro & CellProfiler

- OMERO.webclient
- Browse data
- View images
- Add annotations
-
CellProfiler
- Running headless in Jupyter notebook
- Analyse Data in IDR from RNAi screen: Heriche et al, 2014
- Use CellProfiler Example pipeline: Percent Positive
- Save CellProfiler results as OMERO.table
Session 2: Orbit

- Orbit as an OMERO client, using Java API
- Create a segmentation model in Orbit
- Saved as a File Annotation to OMERO
- Segment an Image, save Polygons as OMERO ROIs
- Use Orbit script editor to access OMERO Java API
Session 3: Fiji, python scripts & R



- Fiji as an OMERO client, using Java API
- Segment images, save Polygons as OMERO ROIs
- Python script on OMERO server summarises ROIs to OMERO.table
- Analyse OMERO.table data in R, plot results
Session 4: OMERO.web: parade & figure
- OMERO.parade: app for filtering and plotting data
- Uses OMERO.table data from Fiji/Python scripts previously
- OMERO.figure: tool for creating figures
- Uses metadata for speed & accuracy
- ROIs, Channel names, Pixel sizes, Tags etc.
Getting started
Importing Images...
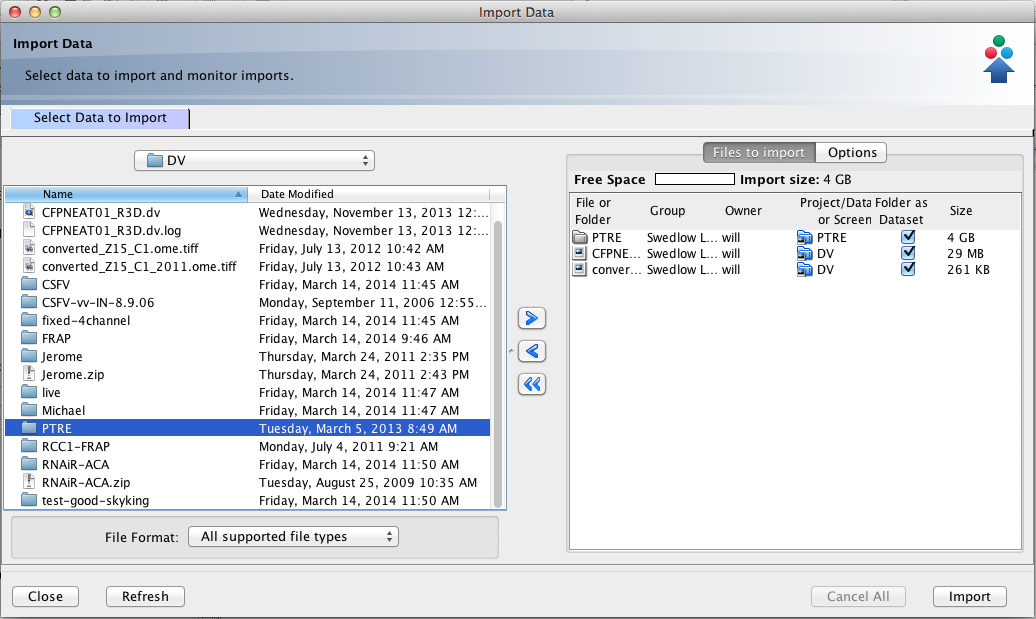
Command-line import
$ bin/omero import image.lsm
In-place import
Files remain in-place during import
# e.g. Transfer file using 'soft' link
$ bin/omero import -- --transfer=ln_s image.lsm