OMERO workshop
Dundee, November 2018
Jean-Marie Burel
Outline
- Intro: OME
- What is OMERO?
- Workshop: Using OMERO
A typical microscopy workflow
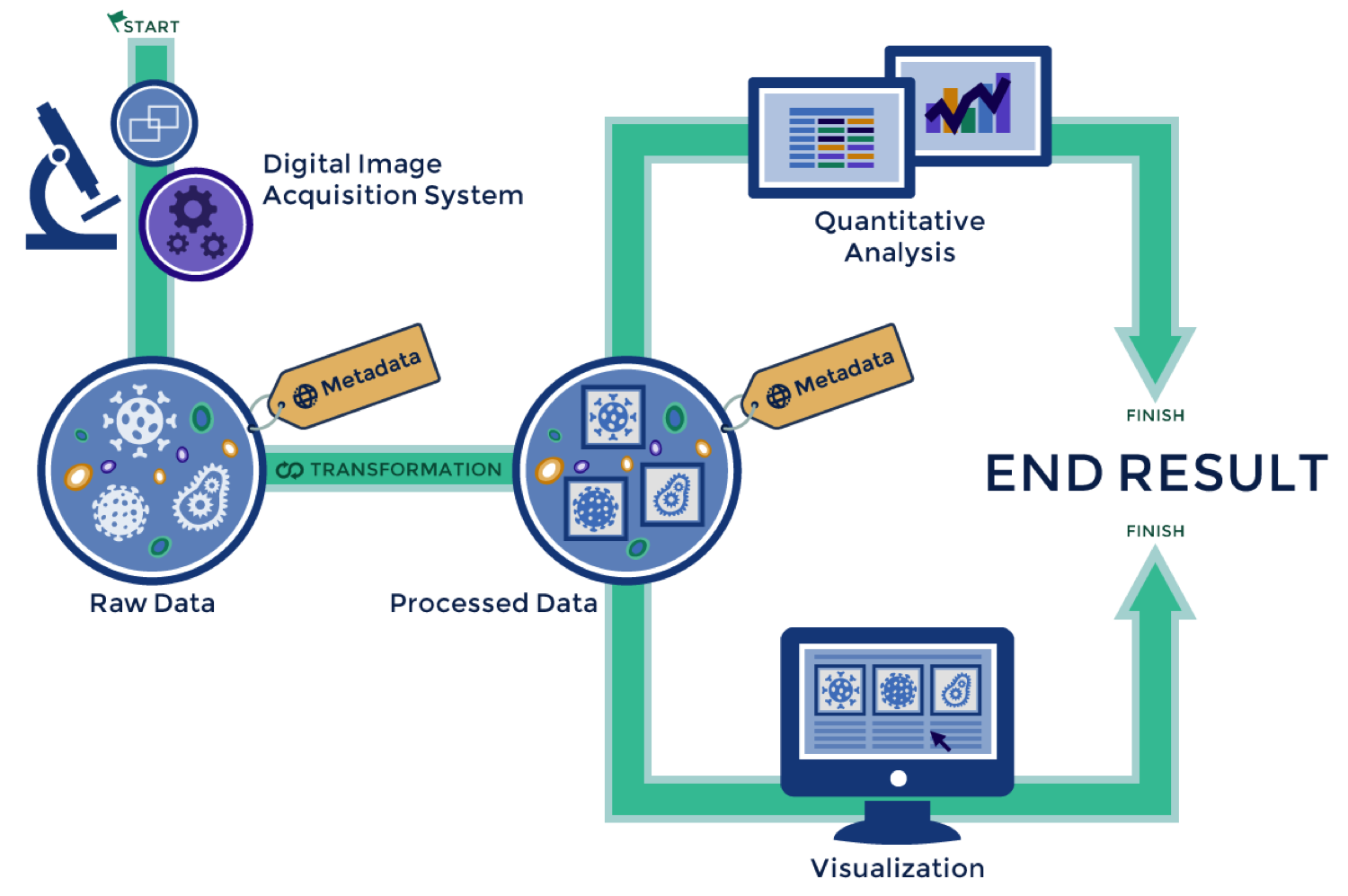
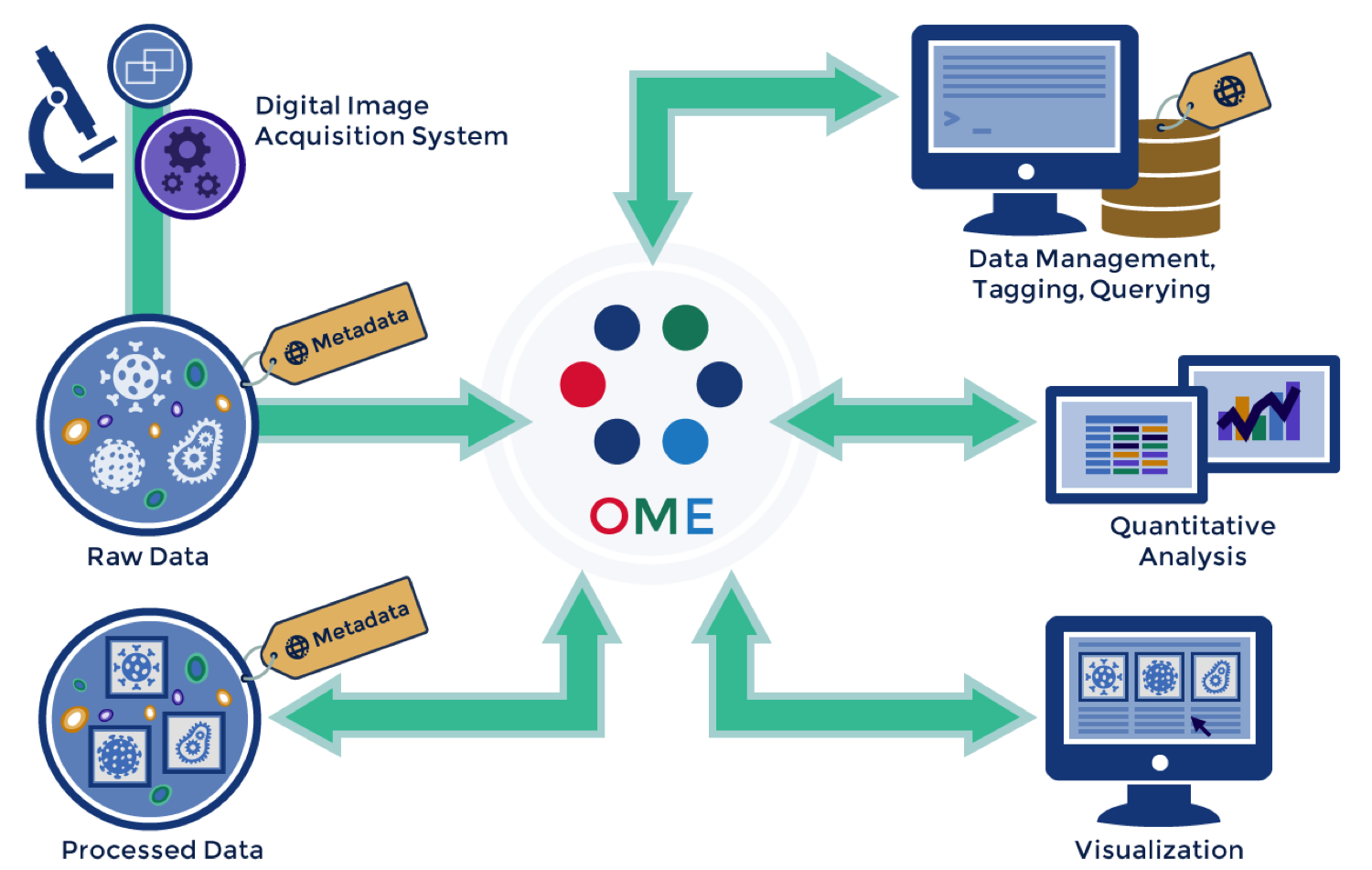
OME: Interoperability
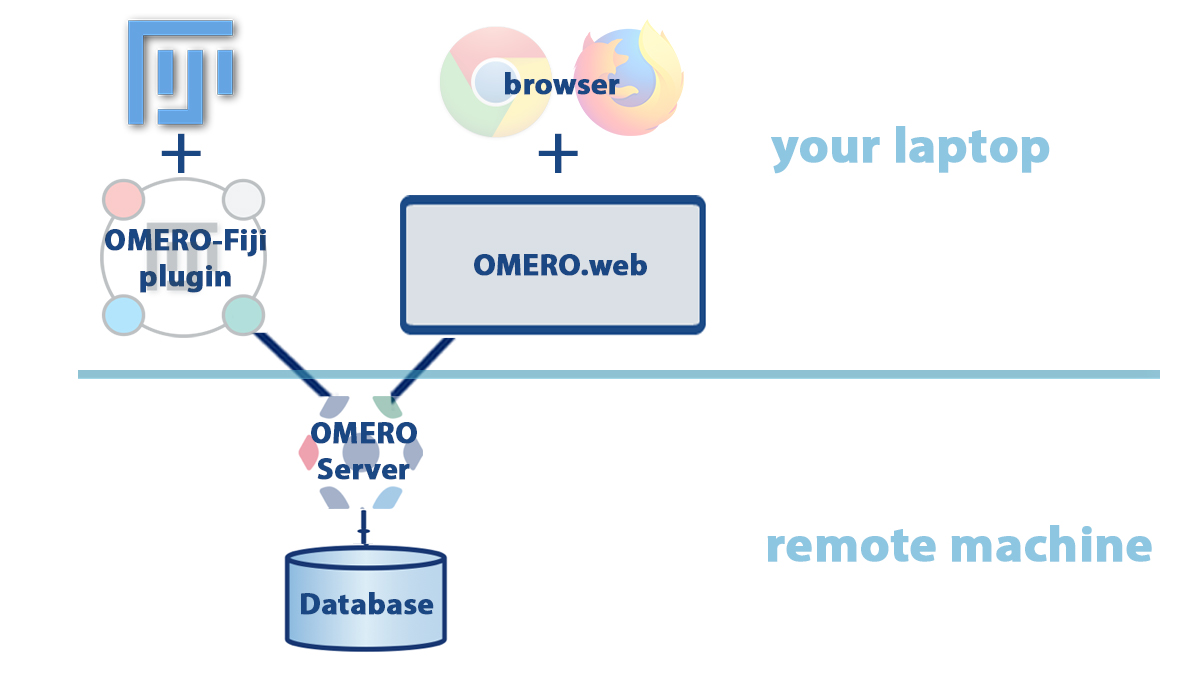
OMERO is a server with clients
Image Data - Read by Bio-Formats
- Support for reading > 150 image formats
- Read pixel data and metadata
- Includes 5D images, HCS data, Tiled WSI images
- Domains: biological, medical, general (tiff, png, etc.)
Metadata
- Stored in a Relational Database
- Acquisition metadata
- User-added Annotations
- ROIs / Segmentations
Search
- Text and Annotations indexed with Lucene
- Allows keyword searches
OMERO.tables
- HDF5 tables for analysis results
- Flexible columns
OMERO API
Python | Java | Matlab | R
OMERO.web framework is extensible
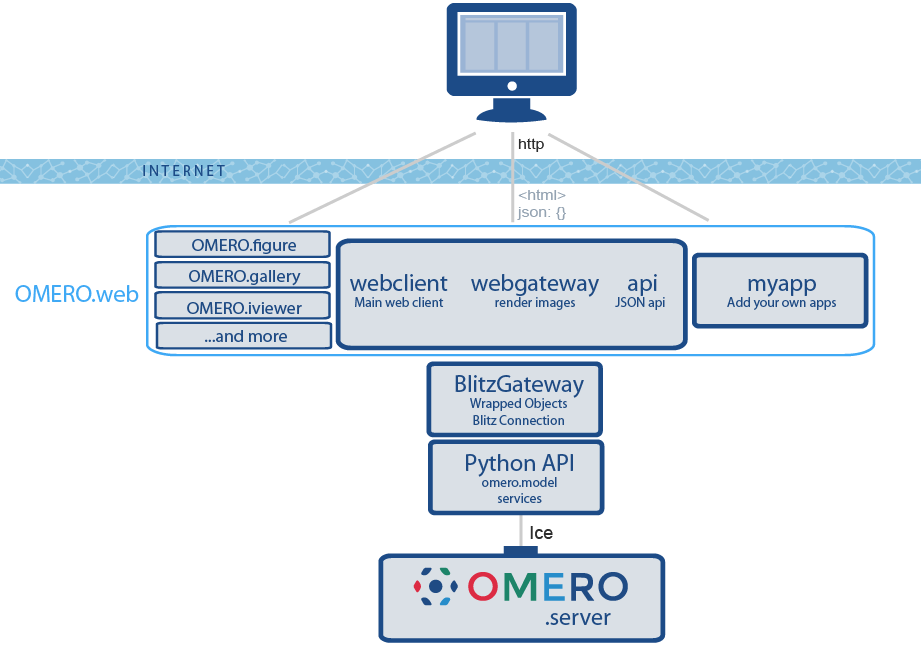
Different clients for different jobs
OMERO clients
- OMERO.insight
- OMERO.webclient
- Browsing images
- Viewing metadata
- Data management
- Searching
- OMERO web apps
- OMERO.iviewer
- OMERO.figure
- OMERO.parade
- ...
- ...
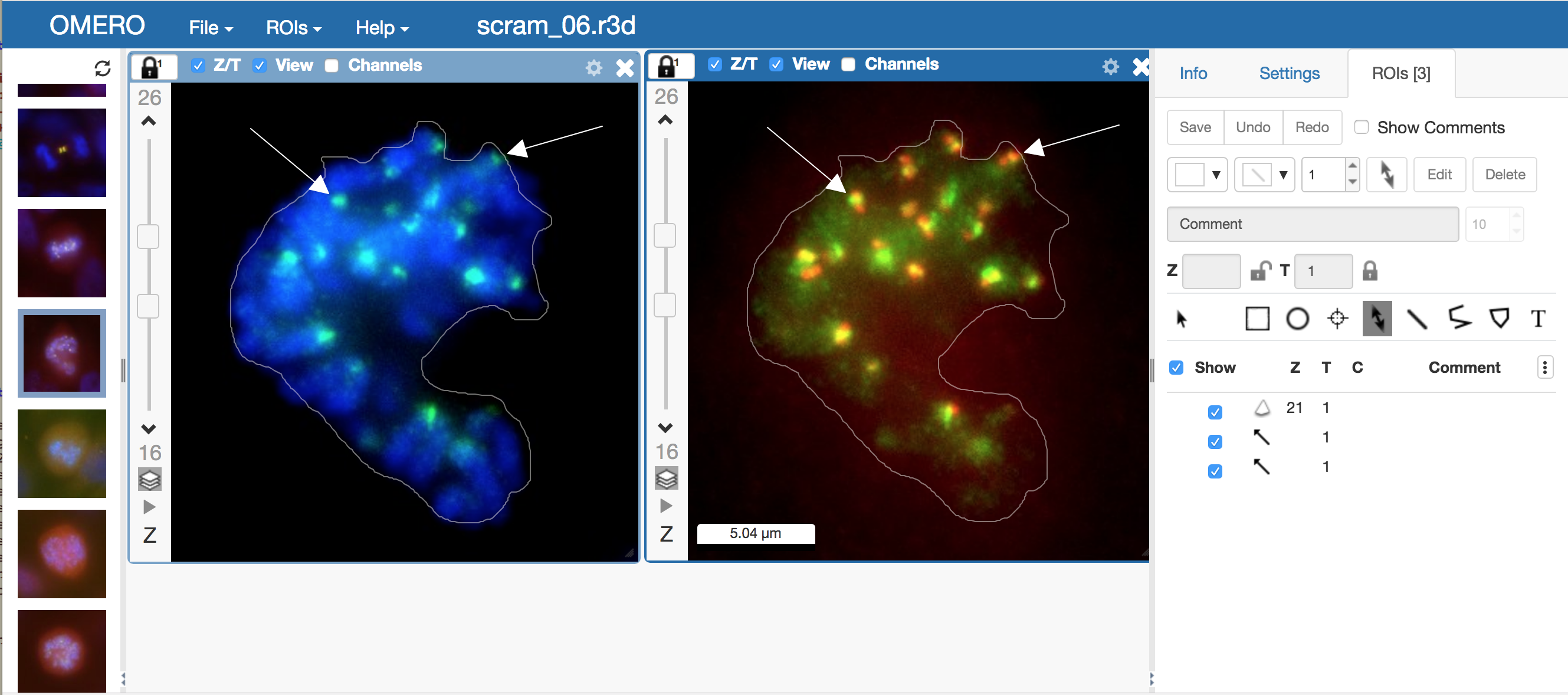
OMERO.iviewer
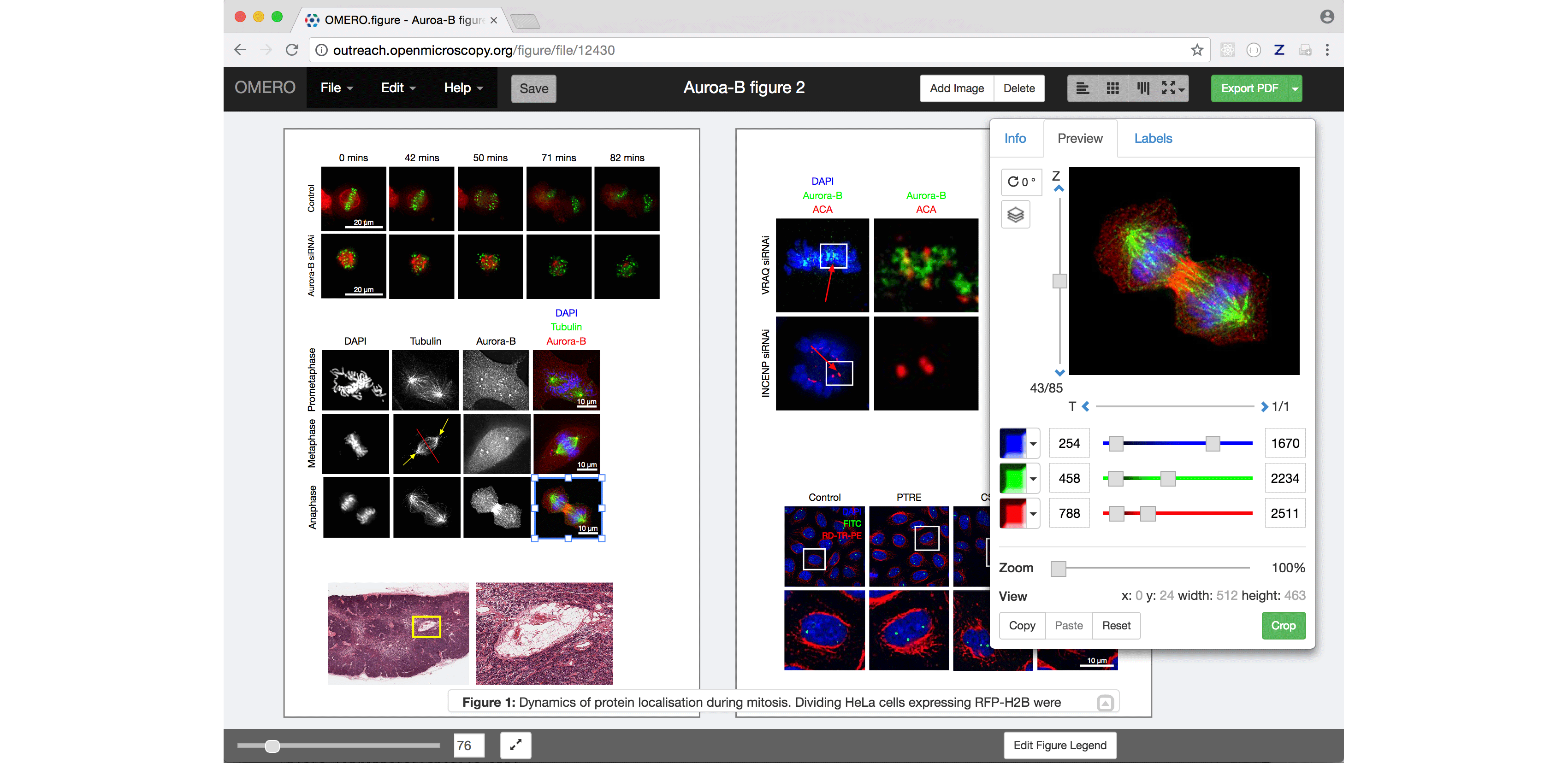
OMERO.figure
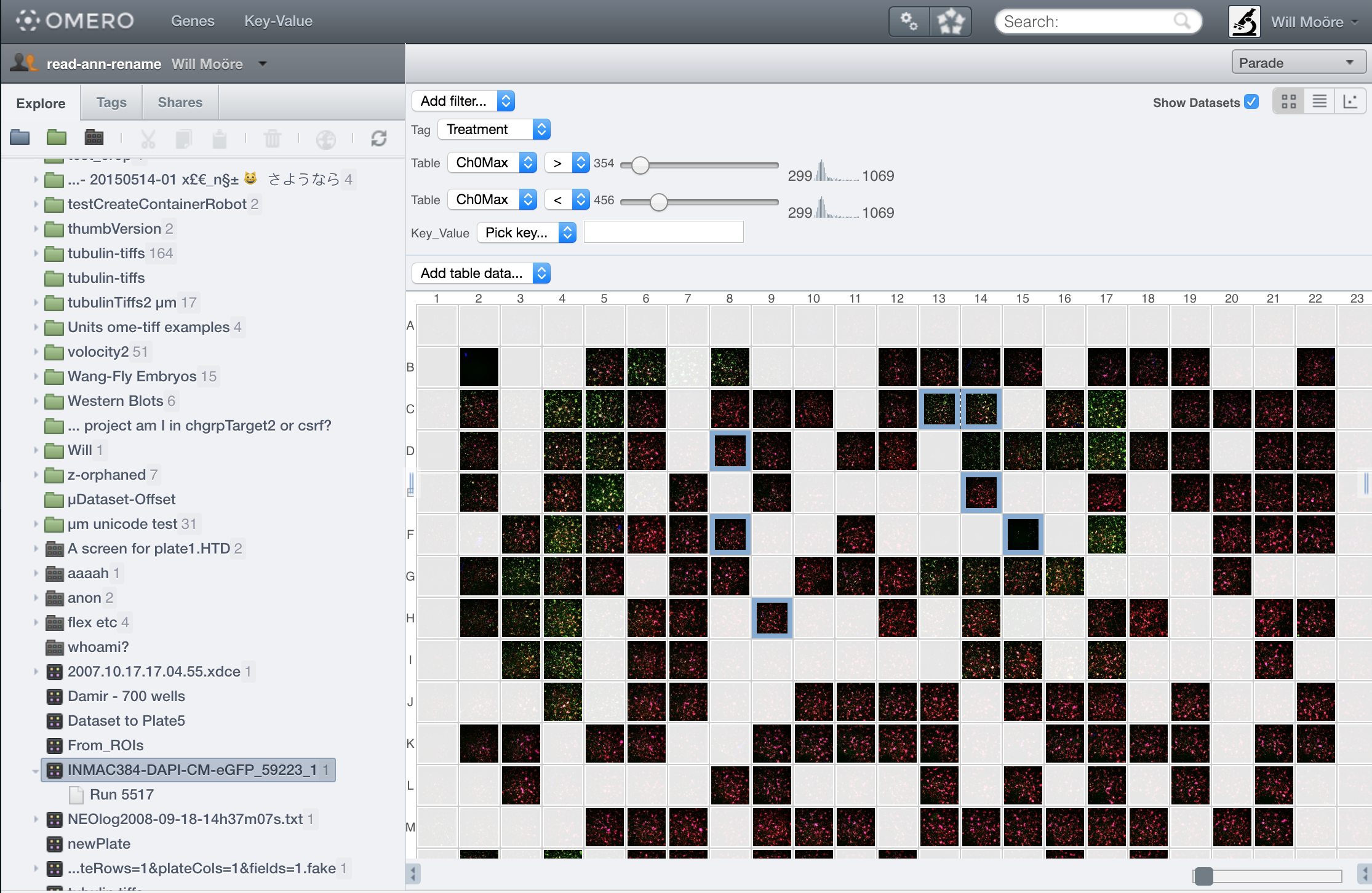
OMERO.parade
Open with... Web apps
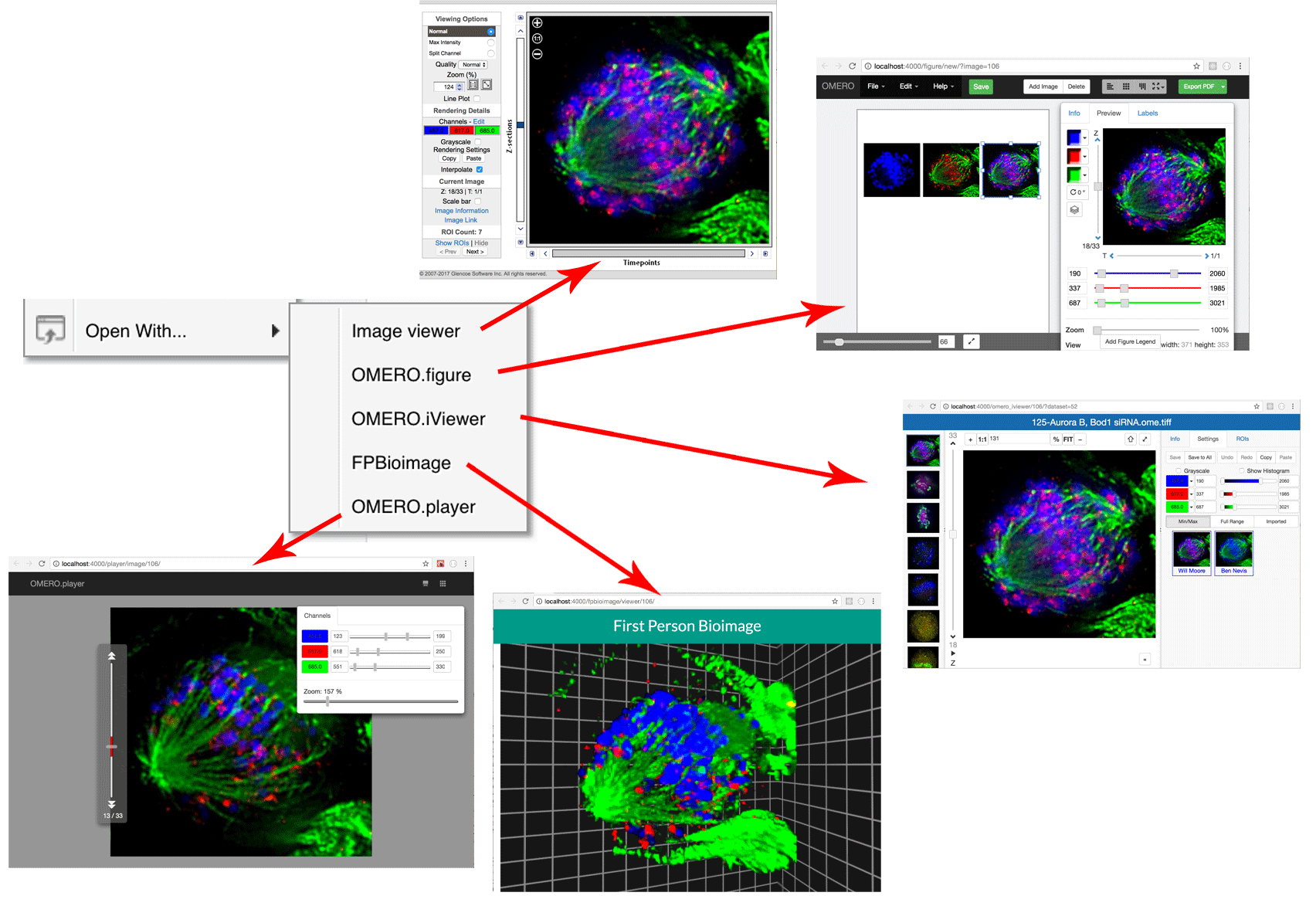
Integration

Set up
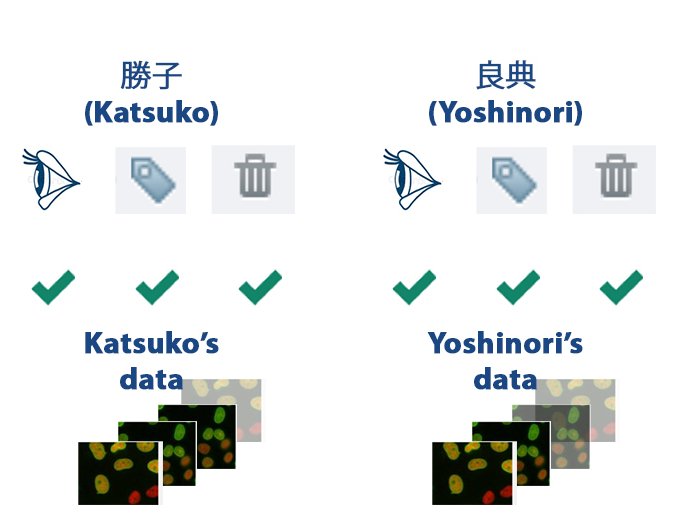
Own Data
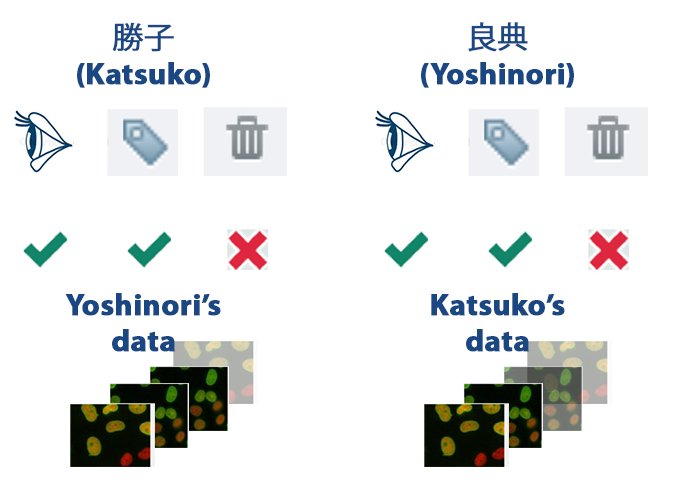
Each others data – read-annotate setup
OMERO in Dundee
- Production server:
- nightshade.openmicroscopy.org
- Free access
- Use LDAP credentials
Thank you







Today's Demo...
outreach.openmicroscopy.org
(Server located in Dundee)