Bio-Formats

- Bio-Formats is a standalone Java library
- It is a community driven project
- Converts proprietary data into an open standard
- Provides a standardized Java application interface
- Supported in numerous applications
- Open source analysis programs like ImageJ, CellProfiler
- Informatics solutions like OMERO, JCB DataViewer
- Commercial programs such as MATLAB
- Can also be used with Python or non Java applications
What can you do now?
Read images and metadata across many domains...
- light microscopy
- electron microscopy
- medical imaging (CT, PET, ...)
- high content screening (HCS)
- digital pathology/whole slide imaging (WSI)
- FLIM
- SPIM
...and in many different applications
Java:
ImageReader reader = new ImageReader();
IMetadata omeMetadata = MetadataTools.createOMEXMLMetadata();
reader.setMetadataStore(omeMetadata);
reader.setId("/PATH/TO/FILE");
for (int plane = 0; plane < reader.getImageCount(); plane++) {
byte[] image = reader.openBytes(plane);
Number timestamp = omeMetadata.getPlaneDeltaT(0, plane).value();
}
ImageJ macro:
run("Bio-Formats Macro Extensions");
Ext.setId("/PATH/TO/FILE");
Ext.getImageCount(imageCount);
timestamps = newArray(imageCount);
for (plane=0; plane < imageCount; plane++) {
Ext.openImage("image #" + plane, plane);
Ext.getPlaneTimingDeltaT(timestamps[plane], plane);
}
MATLAB:
r = bfGetReader("/PATH/TO/FILE");
imageCount = r.getImageCount();
omeMetadata = r.getMetadataStore();
for plane = 1:imageCount
image = bfGetPlane(r, plane, varargin{:});
timestamp = omeMetadata.getPlaneDeltaT(0, plane - 1).value().doubleValue();
end
Python:
reader = bioformats.get_image_reader(None, path="/PATH/TO/FILE")
imageCount = reader.rdr.getImageCount()
omeMetadata = javabridge.JWrapper(reader.rdr.getMetadataStore())
for plane in range(0, imageCount):
image = reader.read(series=0, index=plane, rescale=False)
timestamp = omeMetadata.getPlaneDeltaT(0, plane)
R:
reader = .jnew("loci.formats.ImageReader")
.jcall(reader, , "setId", file)
omeMetadata = .jcall(reader, "", "getMetadataStore")
image = .jcall(reader, "[B", "openBytes", 0)
timestamp = .jcall(omeMetadata, "", "getPlaneDeltaT", 0, 0)
What we've done in the past year
- released 4 new minor versions from 5.2 to 5.5
- new API additions for tiling
- new dynamic metadata options
- added support for 4 new file formats
- release of a new schema 2016-06
- bug fixes and reader improvements
- performance and test coverage improvements
- re architecture of components
New minor releases
See the Bio-Formats version history
- Bio-Formats 5.2 - New schema release, 2 new file formats
- Bio-Formats 5.3 - Tiling API additions, dynamic metadata options, JPEG-XR support
- Bio-Formats 5.4 - command line options, performance improvements
- Bio-Formats 5.5 - Support for Olympus OIR and PerkinElmer Columbus
New file formats & codecs
- Olympus OIR (funded by a partnership between Glencoe Software and OLYMPUS EUROPA SE & Co. KG)
- PerkinElmer Columbus
- Becker & Hickl .spc
- Princeton Instruments .spe
- JPEG-XR compressed CZI data (funded by a partnership between Glencoe Software and ZEISS)
New OME schema - 2016-06
See the Bio-Formats schema history
- introduced the concept of Folders
- ROI properties have been simplified
- extended enum metadata to better support units
- Shape, LightSource and Map are now complexTypes
- MapPairs has been dropped
- Various code generation improvements:
- BinData has been added to handle raw binary data
- simplify and standardize the generation process
- allow for genuine abstract model types
- enable C++ model implementation
Architecture Changes
- decoupled components to new GitHub repositories.
- consumed as artifacts from Maven Central.
- The following components have been decoupled:
- formats-common - now ome-common-java
- ome-poi
- specification, xsd-fu and ome-xml components - now ome-model
- mdbtools - now ome-mdbtools
- stubs components - now ome-stubs
- metakit component - now ome-metakit
Tiling
- New API functions
- Set and retrieve tile size along X & Y
- Tiled writing for TIFF-based formats
- Automatically handles tiled writing
- Can still write tiles using TIFF IFD
Tiling API
- to set up an image writer to use tiling the following 2 API functions are provided:
public int setTileSizeX(int tileSize) throws FormatException
public int setTileSizeY(int tileSize) throws FormatException
Tiling API
- Each function takes in an integer parameter for the desired tile size.
- The image writer will round the requested value to the nearest supported tile size.
- The return value will contain the actual tiling size which will be used by the writer.
Tiling API
- To find out the tiling size currently being used at any point there are 2 further API functions to get the current tile size for a writer.
public int getTileSizeX() throws FormatException
public int getTileSizeY() throws FormatException
Tiled Writing - Introduction
See full example at SimpleTiledWriter
Simple Tiled Writer
// construct the object that stores OME-XML metadata
ServiceFactory factory = new ServiceFactory();
OMEXMLService service = factory.getInstance(OMEXMLService.class);
IMetadata omexml = service.createOMEXMLMetadata();
// setup the reader and associate it with the input file
reader = new ImageReader();
reader.setMetadataStore(omexml);
reader.setId(inputFile);
// setup the writer and associate it with the output file
writer = new OMETiffWriter();
writer.setMetadataRetrieve(omexml);
writer.setInterleaved(reader.isInterleaved());
// set the tile size height and width for writing
this.tileSizeX = writer.setTileSizeX(tileSizeX);
this.tileSizeY = writer.setTileSizeY(tileSizeY);
writer.setId(outputFile);
byte[] buf = new byte[FormatTools.getPlaneSize(reader)];
for (int series=0; series < reader.getSeriesCount(); series++) {
reader.setSeries(series);
writer.setSeries(series);
// convert each image in the current series
for (int image=0; image < reader.getImageCount(); image++) {
// Read tiles from the input file and write them to the output OME-Tiff
// The OME-Tiff Writer will automatically write the images in a tiled format
buf = reader.openBytes(image);
writer.saveBytes(image, buf);
}
}
Tiled Writing - Reading and Writing
See full example at TiledReaderWriter
Tiled Reader Writer
int bpp = FormatTools.getBytesPerPixel(reader.getPixelType());
int tilePlaneSize = tileSizeX * tileSizeY * reader.getRGBChannelCount() * bpp;
byte[] buf = new byte[tilePlaneSize];
for (int series=0; series < reader.getSeriesCount(); series++) {
reader.setSeries(series);
writer.setSeries(series);
// convert each image in the current series
for (int image=0; image < reader.getImageCount(); image++) {
int width = reader.getSizeX();
int height = reader.getSizeY();
// Determined the number of tiles to read and write
int nXTiles = width / tileSizeX;
int nYTiles = height / tileSizeY;
if (nXTiles * tileSizeX != width) nXTiles++;
if (nYTiles * tileSizeY != height) nYTiles++;
for (int y=0; y < nYTiles; y++) {
for (int x=0; x < nXTiles; x++) {
// The x and y coordinates for the current tile
int tileX = x * tileSizeX;
int tileY = y * tileSizeY;
// Read tiles from the input file and write them to the output OME-Tiff
buf = reader.openBytes(image, tileX, tileY, tileSizeX, tileSizeY);
writer.saveBytes(image, buf, tileX, tileY, tileSizeX, tileSizeY);
}
}
}
}
Metadata Options
See full list of Reader and Writer options
- new API supporting arbitrary key/value pairs
- DynamicMetadataOptions class used to set options
- options are now supported in the command line tools
- in FIJI options are available via Bio-Formats Plugins Configuration window
Current Available Options
Reader Options
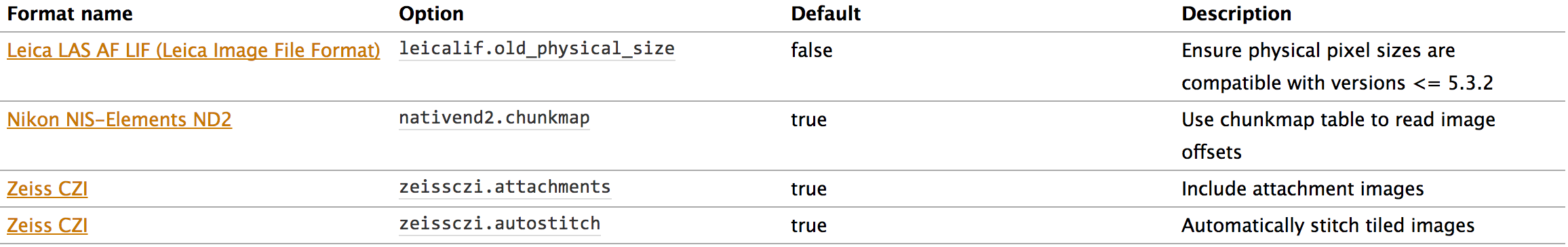
Writer Options

API Usage
Retrieving Options
MetadataOptions options = writer.getMetadataOptions();
if (options instanceof DynamicMetadataOptions) {
boolean value = ((DynamicMetadataOptions) options).getBoolean("ometiff.companion");
}
Setting Options
MetadataOptions options = reader.getMetadataOptions();
if (options instanceof DynamicMetadataOptions) {
((DynamicMetadataOptions) options).setBoolean("nativend2.chunkmap", true);
reader.setMetadataOptions(options);
}
Command Line Usage
- Reader options can be used with showinf -option
showinf example
showinf -option nativend2.chunkmap true inputFile.nd2
- Writer options can be used with bfconvert -option
bfconvert example
bfconvert -option ometiff.companionoutputFile.companion.ome inputFile.tiff outputFile.ome.tiff
What's next?
- Tiling performance benchmarks
- Evaluating new binary containers
- Containing releases of Bio-Formats
- More user contributions
- What else would you like to see?
How can I get involved?
- Contribute on GitHub
- Raise GitHub Issues
- Contact us via mailing lists:
- ome-devel mailing list
- ome-users mailing list
- searchable using google with ‘site:lists.openmicroscopy.org.uk’
- Contact us via the forum
- Submit QA files
Thank you







