OME 2016-06 Data Model
June 14, 2016
Mark Carroll, David Gault, Roger Leigh, Sébastien Besson
Model Changes
- Namespace unification
- Class hierarchies
- ROI Properties
- Folders
Namespace unification
- No more Schemas/SA/2015-01,
Schemas/SPW/2015-01, etc. - For 2016 all now in one OME namespace
Namespace unification
OME-XML impact
2015-01 sample<?xml version="1.0" encoding="UTF-8"?>
<OME xmlns:xsi="http://www.w3.org/2001/XMLSchema-instance"
xmlns="http://www.openmicroscopy.org/Schemas/OME/2015-01"
xmlns:OME="http://www.openmicroscopy.org/Schemas/OME/2015-01"
xmlns:Bin="http://www.openmicroscopy.org/Schemas/BinaryFile/2015-01"
xmlns:SPW="http://www.openmicroscopy.org/Schemas/SPW/2015-01"
xmlns:SA="http://www.openmicroscopy.org/Schemas/SA/2015-01"
xmlns:ROI="http://www.openmicroscopy.org/Schemas/ROI/2015-01"
xsi:schemaLocation="http://www.openmicroscopy.org/Schemas/OME/2015-01
http://www.openmicroscopy.org/Schemas/OME/2015-01/ome.xsd">
2016-06 sample
<?xml version="1.0" encoding="UTF-8"?>
<OME xmlns:xsi="http://www.w3.org/2001/XMLSchema-instance"
xmlns="http://www.openmicroscopy.org/Schemas/OME/2016-16"
xsi:schemaLocation="http://www.openmicroscopy.org/Schemas/OME/2016-16
http://www.openmicroscopy.org/Schemas/OME/2016-16/ome.xsd">
Class Hierarchies
Data representation and code generation simplification
- Elements made complexType
- LightSourceArc, Filament, GenericExcitationSource, Laser, LightEmittingDiode
- ShapeEllipse, Label, Line, Mask, Point, Polygon, Polyline, Rectangle
- MapMapAnnotation, GenericExcitationSource, ImagingEnvironment
- Elements dropped
- MapPairs
- Extended usage of xsd:appinfo for enumerations
Class Hierarchies
Shape
2015-01<ROI ID="3">
<Union>
<Shape ID="Shape:3" FillRule="EvenOdd" Text="Hi There!">
<Rectangle X="1" Y="2" Width="3" Height="4"/>
</Shape>
<Shape ID="Shape:4" FillRule="EvenOdd" FontStyle="Normal" FontFamily="serif">
<Label X="1" Y="1"/>
</Shape>
</Union>
</ROI>
2016-06
<ROI ID="3">
<Union>
<Rectangle ID="Shape:3" FillRule="EvenOdd" Text="Hi There!" X="1" Y="2" Width="3" Height="4"/>
<Label ID="Shape:4" FillRule="EvenOdd" FontStyle="Normal" FontFamily="serif" X="1" Y="1"/>
</Union>
</ROI>
Class Hierarchies
LightSource
2015-01
<Instrument ID="Instrument:0">
<LightSource ID="LightSource:0" Power="200" Manufacturer="OME Lights" Model="Ruby60"
SerialNumber="A654321">
<Laser Type="SolidState" LaserMedium="Ruby">
<Pump ID="LightSource:1"/>
</Laser>
</LightSource>
<LightSource ID="LightSource:1" Power="300" Manufacturer="OME Lights" Model="Arc60"
SerialNumber="A123456">
<Arc Type="Xe"/>
</LightSource>
2016-06
<Instrument ID="Instrument:0">
<Laser ID="LightSource:0" Power="200" PowerUnit="mW" Manufacturer="OME Lights"
Model="Ruby60" SerialNumber="A654321" Type="SolidState" LaserMedium="Ruby" RepetitionRate="1.2"
RepetitionRateUnit="MHz" Wavelength="590.5" WavelengthUnit="nm">
<Pump ID="LightSource:1"/>
</Laser>
<Arc ID="LightSource:1" Power="300" PowerUnit="mW" Manufacturer="OME Lights"
Model="Arc60" SerialNumber="A123456" Type="Xe"/>
Class Hierarchies
MapAnnotation
Bio-Formats 5.1.xm = new MapAnnotation();
List< MapPair > p = new ArrayList< MapPair>();
p.add(new MapPair("a", "1"));
p.add(new MapPair("b", "1"));
m.setValue(new MapPairs(p));
List< MapPair > p2 = m.getValue().getPairs();
Bio-Formats 5.2.xm = new MapAnnotation();
List< MapPair > p = new ArrayList< MapPair>();
p.add(new MapPair("a", "1"));
p.add(new MapPair("b", "1"));
m.setValue(pairs);
List< MapPair > p2 = m.getValue();
Class Hierarchies
ImagingEnvironment
Bio-Formats 5.1.xi = new ImagingEnvironment();
List< MapPair > p = new ArrayList< MapPair>();
p.add(new MapPair("a", "1"));
p.add(new MapPair("b", "1"));
i.setMap(new Map(p));
List< MapPair > p2 = i.getMap().getPairs();
Bio-Formats 5.2.xi = new ImagingEnvironment();
List< MapPair > p = new ArrayList< MapPair>();
p.add(new MapPair("a", "1"));
p.add(new MapPair("b", "1"));
i.setMap(pairs);
List< MapPair > p2 = i.getMap();
Class Hierarchies
GenericExcitationSource
Bio-Formats 5.1.xg = new GenericExcitationSource();
List< MapPair > p = new ArrayList< MapPair>();
p.add(new MapPair("a", "1"));
p.add(new MapPair("b", "1"));
g.setMap(new Map(p));
List< MapPair > p2 = g.getMap().getPairs();
Bio-Formats 5.2.xi = new GenericExcitationSource();
List< MapPair > p = new ArrayList< MapPair>();
p.add(new MapPair("a", "1"));
p.add(new MapPair("b", "1"));
g.setMap(pairs);
List< MapPair > p2 = g.getMap();
ROI Properties
Simplification of graphical aspects of the data model
- Properties dropped
- ROIROI.Namespace
- ShapeShape.LineCap, Shape.Visible
- Marker enumerations droppedCircle, Square
- Arrow markers still available for Line/Polyline
Markers and OMERO
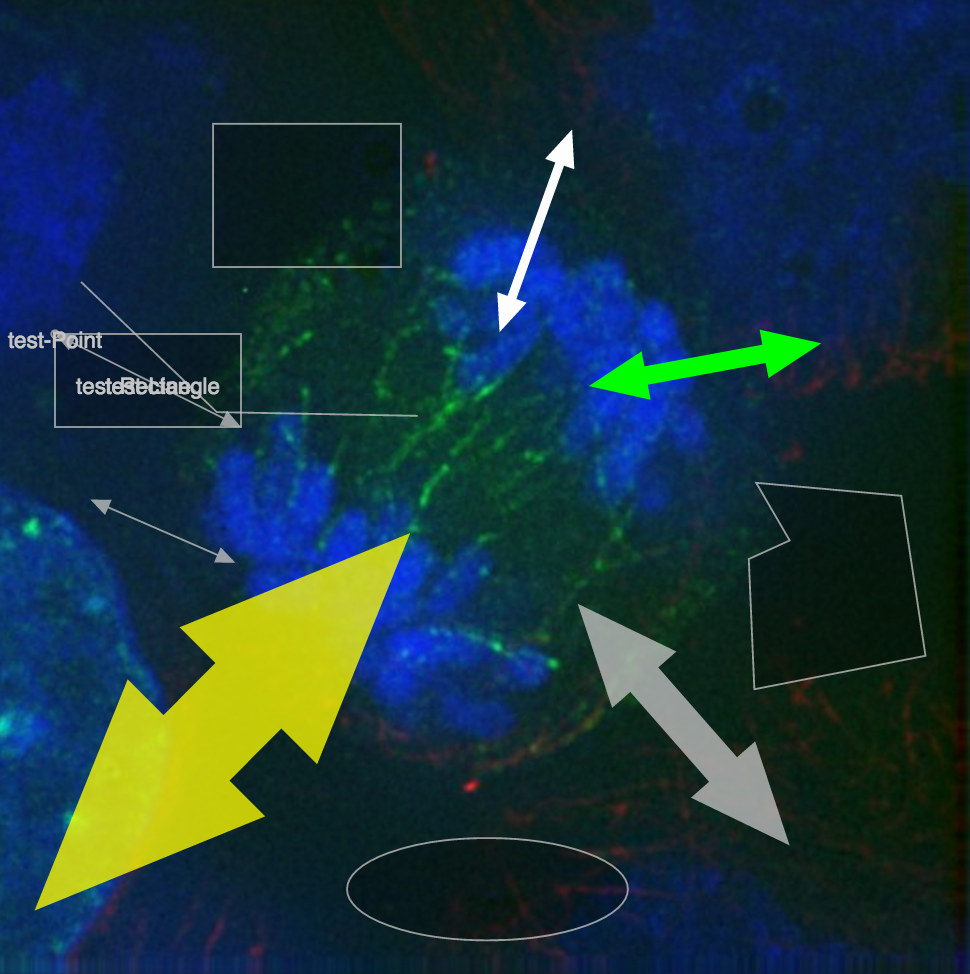
Folders
Allow the organization of ROI elements into hierarchical structures
- Top-level element that may contain: Folders, ROIs, Images
- Folder hierarchy is a strict tree: a Folder may have only one parent
Folder: OME-XML
<Folder ID="Folder:1" Name="CMPO"> <FolderRef ID="Folder:2"/> <FolderRef ID="Folder:7"/> </Folder> <Folder ID="Folder:7" Name="Cell Component"> <FolderRef ID="Folder:8"/> <ROIRef ID="Roi:14"/> </Folder>
Folders and OMERO
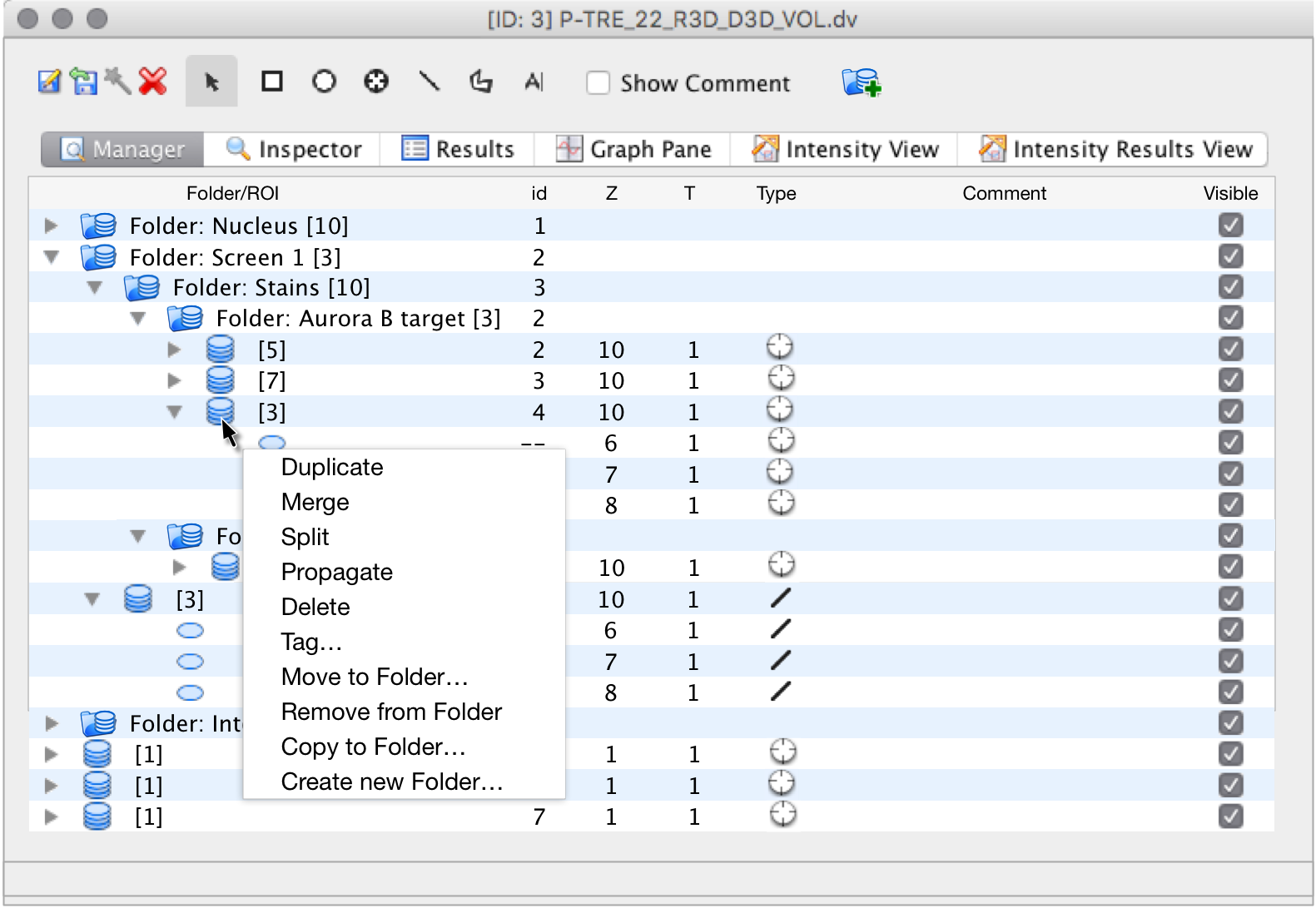
Folders and Images
- Images are associated with Folders,
- directly via ImageRef or
- indirectly via ROIRef
What's next
- June: release of 2016-06 schema
- July: release of OME Files C++ 0.2.0
- August/September: release of Bio-Formats 5.2.0
Thank you
- OME team

