Introduction to Bio-Formats
What is Bio-Formats?
- Java library for reading image-based datasets
- Reads images, but also acquisition metadata
- One API, many applications
- A complete implementation of OME-XML and OME-TIFF
- also now in C++
A brief historical overview
- 2005 - work began on Java library
- 2006 March 31 - first official release of Bio-Formats
- late 2006 - OMERO begins using Bio-Formats
- early 2008 - MATLAB first supported
- 2008 - major effort standardize metadata handling across all formats
- 2009 (v4.0.0) - unified version numbers with OMERO
- late 2012/early 2013 - work began on C++ library
- 2014/2015 - major effort to improve performance
- 2015 - increasing number of contributed code changes
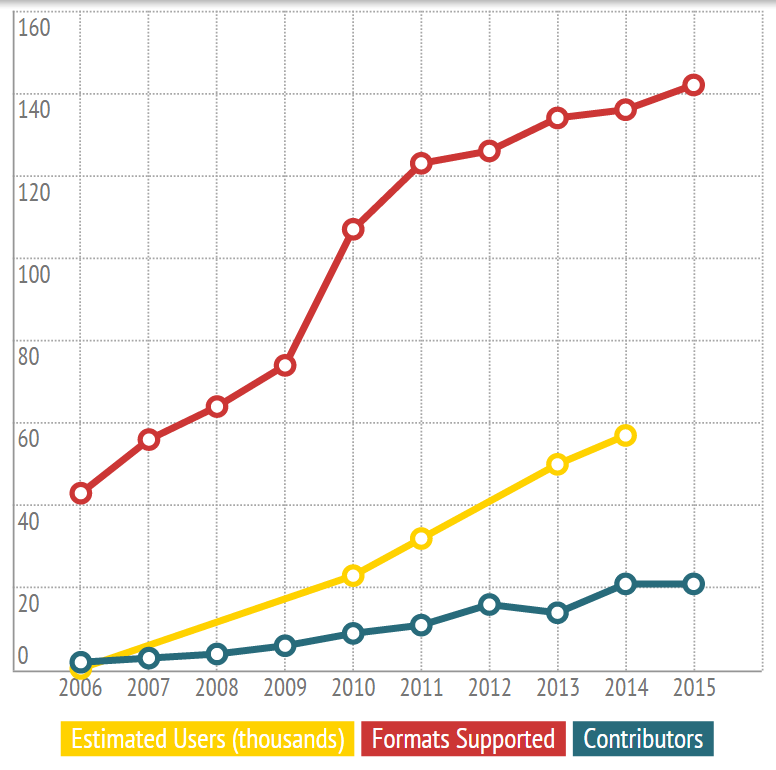
The history in numbers
What can you do now?
Read images and metadata across many domains...
- light microscopy
- electron microscopy
- medical imaging (CT, PET, ...)
- high content screening (HCS)
- digital pathology/whole slide imaging (WSI)
- FLIM
- SPIM
...and in many different applications
Java:
ImageReader reader = new ImageReader();
IMetadata omeMetadata = MetadataTools.createOMEXMLMetadata();
reader.setMetadataStore(omeMetadata);
reader.setId("/PATH/TO/FILE");
for (int plane = 0; plane < reader.getImageCount(); plane++) {
byte[] image = reader.openBytes(plane);
Number timestamp = omeMetadata.getPlaneDeltaT(0, plane).value();
}
ImageJ macro:
run("Bio-Formats Macro Extensions");
Ext.setId("/PATH/TO/FILE");
Ext.getImageCount(imageCount);
timestamps = newArray(imageCount);
for (plane=0; plane < imageCount; plane++) {
Ext.openImage("image #" + plane, plane);
Ext.getPlaneTimingDeltaT(timestamps[plane], plane);
}
MATLAB:
r = bfGetReader("/PATH/TO/FILE");
imageCount = r.getImageCount();
omeMetadata = r.getMetadataStore();
for plane = 1:imageCount
image = bfGetPlane(r, plane, varargin{:});
timestamp = omeMetadata.getPlaneDeltaT(0, plane - 1).value().doubleValue();
end
Python:
reader = bioformats.get_image_reader(None, path="/PATH/TO/FILE")
imageCount = reader.rdr.getImageCount()
omeMetadata = javabridge.JWrapper(reader.rdr.getMetadataStore())
for plane in range(0, imageCount):
image = reader.read(series=0, index=plane, rescale=False)
timestamp = omeMetadata.getPlaneDeltaT(0, plane)
What we've done recently
- full support for Windows
- performance improvements, especially over network file systems
- added units for most metadata values
- lots of bug fixes!
What's next?
- Updates to the development process
- More releases, and faster
- OME-HDF?
- More usage examples and developer documentation
What's next...today?
- Introduction to working with the new C++ library
- Breakouts/technical discussion - look for the Bio-Formats sign!